Abstract
Cells respond to many stressors by senescing, acquiring stable growth arrest, morphologic and metabolic changes, and a proinflammatory senescence-associated secretory phenotype. The heterogeneity of senescent cells (SnCs) and senescence-associated secretory phenotype are vast, yet ill characterized. SnCs have diverse roles in health and disease and are therapeutically targetable, making characterization of SnCs and their detection a priority. The Cellular Senescence Network (SenNet), a National Institutes of Health Common Fund initiative, was established to address this need. The goal of SenNet is to map SnCs across the human lifespan to advance diagnostic and therapeutic approaches to improve human health. State-of-the-art methods will be applied to identify, define and map SnCs in 18 human tissues. A common coordinate framework will integrate data to create four-dimensional SnC atlases. Other key SenNet deliverables include innovative tools and technologies to detect SnCs, new SnC biomarkers and extensive public multi-omics datasets. This Perspective lays out the impetus, goals, approaches and products of SenNet.
Similar content being viewed by others
Main
Senescence is a cell state triggered by numerous cell-intrinsic and cell-extrinsic stressors, including mitotic, oxidative, genotoxic, mechanical or nutrient stress, and organelle dysfunction1. Senescence is driven by p53–p21CIP1 and p16INK4a–Rb tumor suppressor pathways and possibly other signaling mechanisms1,2,3. The senescence response is amplified by several mediators, including ATM, IKK–NF-κB, JAK–STAT, GATA-4 and mTOR. SnCs generally increase in size and protein content, show altered organelle function, chronic genotoxic stress, a robust secretome and resistance to apoptosis1. One common characteristic of SnCs is a stable cell cycle arrest, which prevents a damaged cell from replicating, acquiring mutations and instigating tumorigenesis.
Multiple lines of evidence suggest that SnCs drive aging and diverse age-related diseases in preclinical models1,4,5,6,7,8,9,10. Interventions targeting SnCs impact multiple morbidities of old age11. The senescence-associated secretory phenotype (SASP) includes proinflammatory cytokines, chemokines, growth factors, proteases, receptors, extracellular vesicles, bioactive lipids and extracellular matrix proteases12,13,14. The SASP can drive loss of tissue homeostasis and secondary senescence, but also attract immune cells that mediate tissue regeneration and clear SnCs15. SnCs also have important roles in normal physiology, for example, embryonic development, parturition and wound healing16,17. The tools to discriminate between pathological and physiological SnCs are currently lacking.
In 2011, it was established that genetic clearance of SnCs delays the onset of multiple age-related pathologies in transgenic mice18. In 2016, it was established that genetic clearance of SnCs in mice delays all-cause mortality, extending median not maximum lifespan19, implicating SnCs in many diseases that kill mice, including cancer, chronic kidney disease and cardiomyopathy19. These genetic studies incentivized the development of senotherapeutics—drugs that selectively target SnCs, either killing them (senolytics) or suppressing the SASP (senomorphics). The first senolytics were described in 2015 (ref. 20). Since then, dozens of senotherapeutics have been described, including natural products21,22, repurposed drugs6,23, peptides24, proteolysis-targeted chimeras25 and chimeric antigen receptor T cells26.
Senolytics have proven efficacious in preclinical models of frailty, cardiovascular disease, kidney disease, diabetes, osteoarthritis, osteoporosis, hepatic and pulmonary fibrosis, steatosis, obesity, depression, mortality due to betacoronavirus infection and Alzheimer’s disease27,28. There are numerous ongoing clinical trials testing senolytics in age-related diseases and geriatric syndromes, including frailty, idiopathic pulmonary fibrosis, Alzheimer’s disease, chronic kidney disease, osteoporosis and coronavirus disease 2019. Preliminary data suggest that the senolytic cocktail dasatinib plus quercetin is safe in humans and reduces SnC burden29,30. In mice, a short course of senolytics, administered intermittently, is sufficient to improve multiple measures of physical fitness, even if administered late in life31, highlighting the immense potential impact of senotherapeutics on human health and healthcare costs.
Despite this promise of SnCs as a therapeutic target, there is sparse information about the identity and features of SnCs in human tissues. Little is known about where and when SnCs arise in humans or the extent of SnC and SASP heterogeneity in vivo. Such knowledge could guide therapeutic and organ-specific targeting of SnCs. Clearly, there is a compelling need to develop tools to map and identify human SnCs with spatial and temporal resolution. To address this need, the SenNet Consortium was created in 2021. The goal of SenNet is to functionally characterize the heterogeneity of SnCs in 18 tissues from healthy humans across lifespan at the single-cell resolution, using mice and other models and perturbations for validation. The scientific foundation for this approach is that aging is thought to begin at conception32. Furthermore, SnCs accumulate with chronological age even in the absence of a disease33. Thus, characterizing SnCs across healthy lifespan informs geroscience, the study of fundamental biology of aging enabling identification of therapeutic targets for new interventions that could simultaneously prevent, attenuate and/or delay multiple age-related diseases, for which old age is by far the greatest risk factor34. This approach will also facilitate the discrimination of SnCs that are physiological versus pathological (that is, required for establishing or reattaining tissue homeostasis versus driving or exacerbating disease), while informing future studies of the specific role of SnCs in individual diseases. The approach comes with risks, as there are no reference or control samples. Hence, perturbations that drive or eliminate SnCs will be necessary for confirming SnC identity in healthy tissues.
A key product of SenNet will be multimodal atlases mapping these rare cells in human and mouse organs. Ancillary, albeit impactful, anticipated products will be new technologies to detect, quantify and trace SnCs, as well as biomarkers of SnCs that facilitate translation of senotherapeutics and diagnostics of the numerous chronic diseases in which SnCs have a causal role. To achieve this ambitious goal, several key deliverables are defined for the consortium (see section below). The goal of this Perspective is to define the rationale for SenNet, the approach of the consortium and the anticipated products.
Establishment of SenNet
Key impetuses for aspiring to map SnCs in human tissues are: (1) emerging roles for SnCs in maintaining tissue homeostasis and healing; (2) extensive evidence implicating SnCs in numerous, common age-related diseases, geriatric syndromes and frailty; (3) the advent of a relatively new class of drugs termed senotherapeutics with broad potential applications to improve human health once refined to more-specific SnC targets; and (4) recent advances in single-cell technologies that enable human tissue mapping efforts at unprecedented resolution, making the mapping of SnCs feasible.
To date, human SnCs have largely been characterized in vitro. This research revealed that SnC features depend on the cell type, senescence inducer, temporal dynamics and physiological context. This vast heterogeneity makes it challenging to identify and canonize SnC biomarkers. Thus, no single laboratory, research award or approach can comprehensively define cellular senescence. Yet, this is urgently needed if we are to harness knowledge of SnCs to benefit human health. The tissues, diseases and conditions that affect SnCs during aging and other physiological processes support the need for a community-wide scientific effort. The National Institutes of Health (NIH) Common Fund is a unique and exciting space at NIH specifically designed to address large challenges and opportunities that are of high priority for the entire NIH (all 27 institutes, centers and offices) and biomedical community.
The NIH Common Fund has rapidly mobilized new single-cell and spatial technologies to tackle other large, multidisciplinary and complex biomedical challenges, creating new platforms and tools for the broad scientific community that have tremendous potential to advance human health. The NIH Common Fund is managed by the Office of Strategic Coordination within the Division of Program Coordination, Planning and Strategic Coordination Office of the NIH Director. Common Fund programs must address emerging scientific opportunities and pressing challenges in biomedical research that are transformative, catalytic, synergistic, cross-cutting and unique. Examples of these initiatives include the Human Biomolecular Atlas Program35, and Somatic Cell Genomic Editing36, 4D Nucleome37 and the Genotype-Tissue Expression project38.
In 2021, the NIH Common Fund launched the SenNet program to catalyze the development of a framework for mapping SnCs and their SASP at single-cell resolution in healthy human tissues across development through physiological aging. This comprehensive blueprint for characterizing and mapping SnCs was initiated through several NIH-sponsored workshops engaging internal and external experts working across numerous disciplines. Participants concluded that there is a critical need to develop novel tools and technologies to identify SnCs in vivo and to harmonize data and SnC definitions across laboratories. Model systems and perturbations to validate characteristics of SnCs discovered in tissues were also deemed critical39. For example, mice enable genetic and pharmacologic manipulations of SnCs (production and elimination) as well as longitudinal assessments as organisms chronologically age. In the fall of 2022, SenNet incorporated mechanisms to also establish a murine atlas of SnCs to help to inform the human atlas.
The initial 5-year investment of about $190 million was awarded to eight tissue mapping centers (TMCs), seven technology development and application (TDA) sites and one consortium organization and data coordinating center (CODCC; housed across five sites) to integrate the data developed by the consortium (Fig. 1). The murine effort added five additional TMCs and five TDA sites. SenNet is purposely designed to have a single CODCC to harmonize and integrate efforts from all sites and awardees to create atlases of SnCs that capture information about the evolution of senescence in space and time (four-dimensional (4D) atlases) across the human life course. Eighteen tissues are currently covered by SenNet (Fig. 2).
TMCs are responsible for all aspects of data generation from tissue collection and analysis to data integration and interpretation. We anticipate that TMCs will acquire and integrate imaging and omics data to benchmark, standardize and validate SnC maps at single-cell resolution for their assigned tissues. The TDA sites are responsible for development of innovative, new approaches and tools necessary to deeply phenotype SnCs in human tissues and model systems. Examples include multi-omics characterization of the 4D nucleome in SnCs, high-throughput quantification of telomere-associated foci, and in vivo detection of SnCs via positron emission tomography imaging. Once developed, these new technologies are expected to be applied broadly and collaboratively across multiple tissues by the TMCs. The CODCC will collect, store and curate all data and metadata generated by the TMCs and TDA sites. The CODCC is responsible for generating the computational models, and final atlas products as well as the tools to visualize and disseminate the data as a resource for the broad scientific community
It is expected that SenNet will interface with other cell mapping programs such as Human Bimolecular Atlas Program (HuBMAP), Human Cell Atlas (HCA) and the Kidney Precision Medicine Project (KPMP). HuBMAP is an NIH Common Fund Initiative to develop the resources and framework to map the >30 trillion cells that make up the human body using protein identifiers of cell lineage. HCA is using single-cell and spatial transcriptomics to create cell reference maps defining the position, function and characteristics of all cells in the human body. The KPMP is an initiative of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) aimed at using state-of-the-art and emerging technologies to characterize renal biopsies from participants with acute kidney injury or chronic kidney disease to enable personalized approaches to their treatment. Interfacing with these existing (and future) cell mapping initiatives will save extraordinary amounts of time and money (for example, using antigens and antibodies validated by HuBMAP to identify SnC lineage). It will also add immense value if datasets use common descriptors to define cell subsets and the datasets can be integrated (for example, information on renal SnCs can be integrated with KPMP data on diseased tissue to advance diagnostic and therapeutic approaches). Ideally, the atlases generated by SenNet can be ‘layered’ on top of atlases of cell types in organs in healthy and diseased tissues generated by other mapping programs. Hence, structurally, the SenNet CODCC is similar to and cross-pollinates other NIH-sponsored cell mapping initiatives.
Characterization of senescent cells
Currently, no SnC-specific biomarker exists. Hence, the first goal of SenNet is to explore and define SnC biology. To address this challenge, SenNet created a Biomarker Working Group, which is responsible for iteratively compiling lists of cell traits, RNAs, proteins, lipids and metabolites that may be used to identify SnCs. This will enable modeling and annotating SnC classes and possibly subclasses that are anticipated to be dictated by cell origin, senescence inducer, tissue environment and human age. The overall goal of the Biomarker Working Group is to curate a database of senescence-associated biomarkers. The short-term goal is to generate a list of senescence-associated biomarkers currently used by members of the SenNet Consortium. The information collected will include cell type, the combinations in which biomarkers occur within a single cell, reagents used for detection (for example, antibodies or nucleic acid probes) and their compatibility with various experimental approaches and human tissues. The list is expected to evolve over time, with some markers being removed owing to lack of specificity or sensitivity, and others being added as our understanding of the senescent phenotype improves. As data collection ramps up, artificial intelligence will undoubtedly be crucial for refining the biomarker list. Ultimately, the Biomarker Working Group will produce a compendium of senescence biomarkers at the tissue and cell-type level. Our prediction is that multiple overlapping, non-static signatures of SnCs will ultimately be identified that require detection and quantification of more than one type of biomolecule, making it challenging to detect SnCs by a single method.
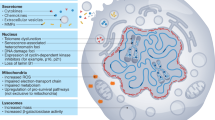
The complexity of senescence entails kinetic alterations in almost all aspects of cell biology, from epigenetic remodeling40 to changes in the quantity and function of organelles41. In vitro studies of oncogene-induced senescence, replication-induced senescence and genotoxin-induced senescence revealed several, frequently generalizable characteristics of SnCs. To date, three main phenotypes characterize SnCs, with the caveat that they are context dependent. Generally, SnCs (1) enter an essentially permanent arrest of proliferation; (2) become relatively resistant to cell death; and (3) develop the SASP. Current biomarkers used to identify SnCs include increased expression of the cell cycle regulators p16INK4a (ref. 42) and p21CIP1 (ref. 43), increased lysosomal senescence-associated β-galactosidase activity44, decreased lamin B1 (ref. 45), increased secretion of HMGB1 (ref. 46) and several markers of genotoxic stress including senescence-associated DNA damage foci of γH2AX and 53BP1, telomere-associated or telomere dysfunction-induced foci characterized by DNA damage response foci at telomeres47, senescence-associated heterochromatic foci characterized by colocalization of dense DAPI staining and modified histones, and senescence-associated distensions of satellite DNA characterized by CENP-B foci at centromeres. Ideally, an endpoint associated with each of the three main phenotypes should be measured to determine if a cell is senescent. Relying on a single endpoint is fraught with error. For example, high senescence-associated β-galactosidase activity is detected in cultured confluent fibroblasts48,49 and certain activated macrophages50,51, whereas p16INK4a and p21CIP1 expression can be induced in a reversible manner under certain physiological contexts50,51,52,53,54.
In addition to the above SnC biomarkers, activation of LINE-1 retrotransposable elements55,56, cytoplasmic chromatin fragments57 and mitochondrial DNA58 are detected in SnCs. Numerous other molecules are attributed to SnCs or the SASP. However, in the absence of cross-validation with established SnC biomarkers at single-cell resolution and validation with appropriate perturbations provoking or targeting SnCs, these molecules are only potential biomarkers of SnCs. As more cell types and physiological contexts are studied, a universal senescence-specific marker may never emerge. Regardless, deep characterization and localization of SnCs in vivo will advance options for diagnosis and treatment of multiple diseases of old age. Internal and external collaborations are an important part of SenNet to facilitate adaptation of emerging technologies and cross-validation. Hence, it is difficult to define the entire scope of features that will ultimately constitute SnC signatures.
Key challenges lie ahead. As stated, none of the current SnC biomarkers are specific to SnCs, requiring multimodal measurement of multiple endpoints at the single-cell level just to identify SnCs1,59, let alone characterize them further. Most published studies rely on bulk tissue analysis or, if at single-cell resolution, implement one method to measure one type of biomolecule. Neither approach is adequate to precisely identify SnCs in tissues, let alone their lineage, their unique characteristics and to predict their role in physiological aging. Multiple targeted and unbiased approaches are required (Fig. 3 and see Supplementary Information for method details) and integration of multi-omics data will be necessary to achieve the goals set forth by SenNet. Considerable value is added by comparing and contrasting characteristics of SnCs of similar or distinct lineages across tissues to discover common (for example, increased expression of a cell cycle inhibitor) and unique (for example, increased expression of a particular SASP protein) SnC features as potential biomarkers and molecular targets. This emphasizes the need for a trans-NIH effort and justifies the structure of SenNet (for example, multi-site, multi-platform, technology development, and a single-data integration site).
CyTOF, cytometry by time-of-flight; scCITE-seq, cellular indexing of transcriptomes and epitopes by sequencing; sc/snRNA-seq, single-cell or single-nucleus RNA sequencing; snATAC-seq, single-nucleus assay for transposase-accessible chromatin using sequencing; MINA, multiplexed imaging of nucleome architectures; IMC, imaging mass cytometry; CODEX, co-detection by indexing immunofluorescence; DBiT-seq, deterministic barcoding in tissue for spatial-omics sequencing for co-mapping mRNAs and proteins; RNAScope, RNA in situ hybridization visualization of single molecules; MERFISH, multiplexed error-robust fluorescence in situ hybridization; GeoMx, NanoString GeoMx digital spatial profiling; Visium, Visium 10x Genomics molecular profiling; Seq-Scope, a spatial barcoding technology with spatial resolution comparable to optical microscopy; Pixel-seq, polony-indexed library sequencing.
Senescent cell atlas
Creating a multiorgan 4D atlas of SnCs with healthy human aging will yield an important tool for investigating disease mechanisms relevant to the mission of most NIH institutes and centers. Currently, we have no knowledge as to whether different types of SnCs appear with advancing physiological age, and/or if SnC phenotypes evolve over time in vivo. Another possibility is that SnCs arise specifically because of acute tissue injury or disease but immune clearance of SnCs declines with age, precipitating chronic disease. Indeed, preexisting SnCs impair host responses to tissue injury or infection5, thereby promoting disease in a feed-forward mechanism.
Single-cell technologies for imaging and deep phenotyping of SnCs have tremendous clinical and translational potential. Complementary, multimodal characterization of SnCs will not only deepen our understanding of senescence biology in health but also reveal the clinical significance of SnCs in cancer, fibrosis, metabolic disorders and diverse degenerative diseases. As bioinformatics approaches on multi-omics are evolving, it will be possible to integrate all epigenomics data with cellular composition for identification of SnC phenotypes. The SenNet Consortium is geared toward deconvoluting the cellular senescence phenotypes based on bioinformatic multi-omics approaches. Although the current goal of SenNet is the mapping of SnCs in ‘normal/healthy’ human and murine tissues to generate reference atlases of SnCs, we anticipate future efforts will leverage these data to study the role of SnCs in various human pathologies.
Given the multiorgan and multimodal data generation envisioned, a structured, cross-team data management, organization and analysis plan is essential. The SenNet CODCC will manage data curation, integration, analysis, atlas creation and dissemination through the SenNet Data Portal (Fig. 4). These harmonization and integration efforts will be coordinated with the Common Fund Data Ecosystem to align SenNet for integration with data from other Common Fund programs. Uniformly processed molecular and cellular data will be integrated with the common coordinate framework (CCF) and will be the basis for construction of an atlas of SnCs. To facilitate uniform data processing and quality-control pipelines within CODCC, and reuse by other data consumers, CODCC will mandate data submission using common data formats that are aligned with CCF reference atlas construction. Examples are the use of Azimuth for cell-type annotation or validated organ mapping antibody panels (OMAP). Uniform processing pipelines will implement state-of-the-art algorithms for the analysis of imaging, sequencing and multi-omics, which will generate standardized datasets that are spatially registered, segmented and annotated using CCF 'Anatomical Structures, Cell Types and Biomarkers’ (ASCT + B) terminology and hence linked to existing ontologies. Integrated and harmonized datasets will be made available through the data portal, along with the raw data.
CCF exploration user interface (EUI) and Vitessce (a visual integration tool for exploring spatial single-cell datasets) will be integrated to enable seamless navigation across scales and queries of SenNet data. The CCF EUI enables registered tissue blocks from the registration user interface (RUI) to be explored spatially (via body browser in the left screenshot, center) and using ontology terms (via hierarchy in the left screenshot, on left) at anatomic scale. A click on a tissue dataset (left) leads to Vitessce (right), which supports the exploration of cellular and molecular scale distributions. EUI provides clinical and spatial context and ontology cross-links, while Vitessce supports details on demand at the molecular scale.
The SenNet Data Portal will also integrate the CCF Registration User Interface (CCF RUI), CCF Explorer User Interface (CCF EUI) and the Vitessce framework in support of exploratory visualization of existing data across levels—from the whole body to single organs to molecular-level and cellular-level datasets and vice versa (Fig. 4). Clinical data will also be standardized and shared in an extension of the CODCC and CCF efforts and will be the basis for standardized implementation and association with electronic health record clinical data in the future.
The CCF consists of ontologies, libraries and computer-based and other training materials that support the efficient mapping, registration and exploration of clinically, semantically and spatially indexed human tissue data. SenNet will extend the HuBMAP CCF that consists of: (1) a CCF Specimen Ontology, which provides CCF-relevant demographic and clinical metadata about the specimen and donor (the ‘who’); (2) a CCF Biological Structure Ontology, which describes ‘what’ part of the body a tissue sample came from; and (3) a CCF Spatial Ontology, which indicates ‘where’ the tissue is in a three-dimensional (3D) reference system. In addition, the CCF defines a ‘registration process’ that makes it possible to annotate data and map it to the 3D reference system, as well as an ‘exploration process’, which facilitates query, analysis and visual examination of registered tissue data and prediction of properties (for example, what cell types are commonly located in a specific anatomical structure or what antibodies should be used to identify a desired set of protein biomarkers) (Fig. 4).
The CCF also provides 3D representations of anatomy linked to ASCT + B tables60. Note that the CCF is semantically explicit (that is, terminology for anatomical structures, cell types and biomarkers link to existing ontologies, namely Uberon/Foundation Model of Anatomy, Cell Ontology (CL) and HUGO Gene Nomenclature Committee) as well as spatially explicit (for example, 3D reference organs are used for registration and exploration). In February 2022, there were ASCT + B tables for 25 organs and 50 associated 3D reference object sets (1–4 per organ, for example, 1 uterus but 4 kidneys to capture left–right and male–female versions), which represent the size, shape, position and spatial orientation of major anatomical structures in an organ-specific manner. The ASCT + B tables and associated spatial reference objects represent the human body in a simplified manner as a partonomy where each cell is part of an anatomical structure that is part of larger anatomical structures and ultimately makes up the entire body.
The SenNet CCF Atlas and SenNet CODCC Data Portal will serve as the ‘hub’ for data coordination and integration. Future extensions of the CCF will require integrating specimen ontology with clinical informatics and electronic health record-based clinical data to characterize not only the state of the participant when the biospecimen was acquired, but also the evolution of the person over their entire lifetime. Furthermore, this may serve as an integration point for environmental factors or cumulative drug exposures. Such examples may then be used to interpret an individual’s ‘health’ atlas using artificial intelligence platforms.
Challenges to creating a 4D SnC atlas include: (1) SnCs are rare in vivo; (2) spatial-omics is a nascent technology implying an additional burden of validation for ill-characterized cell types such as SnCs; (3) for any single SnC biomarker, it is not yet established whether changes in mRNA, protein or the epigenome (or a combination) best reflect a senescent state; (4) implementing a biomarker panel that includes a combination of proteins, nucleic acids, morphology markers and measure of enzymatic activity endpoints currently limits the ability to colocalize SnC biomarkers at single-cell resolution; (5) SnCs in different tissues will probably express common as well as tissue-specific patterns of senescence features; and (6) a lack of tools to confidently discriminate pathological versus physiological SnCs. In complex tissues, both the physiological and pathological roles of SnCs may occur in close proximity (for example, chronic tissue damage foci with adjacent areas of tissue regeneration). To optimize senotherapeutics and minimize side effects of this new class of drugs, one would like to distinguish between SnCs involved in these two processes and to do so using a biomarker measured in an easily accessed tissue or biofluid. This will require tissue mapping advances as well as biomarker discovery in human biofluids.
SenNet deliverables
To produce comprehensive and high-resolution atlases of SnCs, several key SenNet deliverables are anticipated (Fig. 5). First, production of extensive multi-omics and imaging datasets that functionally and spatially identify and characterize SnCs at the single-cell level in 18 human tissues across the life course of humans. The datasets will be made readily accessible to the broad scientific community and searchable. Innovative visualization tools will be developed to maximize the value and accessibility of the data. Second, mapping rare and heterogeneous SnCs in human tissues will require the generation of new tools, technologies and computation modeling systems. Third, the data will yield biomarker panels that enable the identification of SnCs, define their secretome, and illustrate the common principles and heterogeneity of SnCs in the human body. Fourth, validation of SnC biomarkers will require establishing reliable approaches for perturbing SnCs (eliminating, modifying and removing SnCs). Finally, improved imaging tools will be needed to rigorously identify SnCs and their unique properties in vivo, with the aspirational goal of ultimately being able to do so longitudinally at the whole-organism level. The ability to detect SnCs noninvasively and longitudinally in people would substantially improve our ability to monitor the effects of injury, inflammation, carcinogenesis, autoimmunity and responsiveness to specific drugs or biologics, ultimately identifying those who may benefit from senotherapies.
SnC atlas building requires a framework for layering data. Data generated by the TMCs and TDA sites are input into the CODCC along with associated metadata. The datasets are organized and de-identified (curation), then analyzed and integrated. The goal is to create an atlas and public database of curated data that can be searched, analyzed and visualized as 3D images of organs using unified annotations. High-quality experimental data are needed to create a human reference atlas. The evolving reference atlas supports data standardization and federation, making it possible to integrate data from different specimens, laboratories and assay types. The atlas characterizes the healthy human—from the whole body down to the single-cell level; it can be compared across ages and diseases to understand differences, advance research and improve human health. Use case scenarios for different stakeholders (researchers, practitioners and students) guide atlas construction and usage but also experimental data acquisition and analysis. Of note, diversity in terms of human participant gender, race and socioeconomic status is emphasized in SenNet. However, these variables may impact SnC heterogeneity even further, meaning that, in the timeframe of the initial grants, statistically meaningful characterization of SnCs across diverse populations might not be achieved.
A clear and comprehensive definition of SnCs in multiple organs will enable identification of molecular targets unique or enriched in SnCs that could form the basis of selective senotherapeutics to advance the treatment of senescence-related pathologies. Biomarkers will ideally be validated within and across tissues, ultimately enabling predictive modeling, optimizing SnC targeting and ensuring the safety and efficacy of senotherapeutics. A deeper, temporal understanding of SnCs with physiological aging will enable the development of therapies that promote the beneficial effects of SnCs while suppressing or removing the deleterious effects.
The timelines for the development of these key deliverables are as follows. During year 1 (mid-2021 to mid-2022), the consortium will establish policies and guidelines to facilitate collaboration, harmonization and rigor of SenNet activities. Working groups will be established to inform consortium-wide activities and facilitate interfacing with other cell mapping initiatives. These include working groups on policy, benchmarking, biomarkers, omics mapping, imaging, data submission, CCF, Common Fund data ecosystem integration, publication and outreach. The working groups reflect every aspect of the SenNet project pipeline from setting standards for high-quality data generation, annotation and integration in a standardized format, to data dissemination and visualization. In years 2–5 of the consortium (mid-2022 to mid-2026), TMCs are expected to regularly generate large volumes of multi-omics and imaging data, and to develop two-dimensional maps of SnCs in human and mouse tissues. This will require collaborations between TMCs, and with TDA sites, to validate detection tools and methods, and to determine the extent of variability in SnC abundance and features between tissues and individuals across the aging process. As data are generated, the CODCC will create a framework for depositing and visualizing the data in four dimensions. It is expected that the CODCC will release datasets regularly for peer review, publication and sharing. As an example, the first release of data from HubMAP comprised data and metadata from de-identified donors for seven tissues. This included >300 datasets defining the tissue samples and data generated from them via microscopy, mass spectrometry, sequencing and other modalities. Future data releases are scheduled biannually.
Concluding remarks
SenNet’s vision is to identify and functionally characterize the heterogeneity of SnCs across numerous human tissues at single-cell resolution, from embryonic development through to physiological aging. Novel technologies and tools will be created and applied to the characterization of human SnCs. Perturbations and studies in mice will be used to validate new SnC biomarkers. Generating SnC atlases via a collaborative consortium, integrating the data through a single-data coordinating center, and harmonizing this effort with other cell mapping initiatives will amplify the value of SenNet. This NIH Common Fund effort will undoubtedly pave the way for exciting, new possibilities in understanding and therapeutically modulating senescence-associated human conditions.
Future perspectives
SenNet will serve as a unique and comprehensive resource to elucidate the heterogeneity of SnCs elicited in different cell types, by different drivers of senescence, anatomical location and human age. Ideally, novel biomarkers to identify SnCs and distinct signatures of disease-specific and beneficial SnCs will be discovered. The knowledge gained can be deployed to better understand the role of SnCs in health and disease, and to guide clinical translation of senotherapeutics. New biomarkers of SnCs are anticipated to be valuable for identifying individuals at risk of disease, stratifying participants in interventional studies, monitoring the response to senotherapeutics, evaluating therapeutic efficacy and ultimately optimizing and personalizing interventions.
References
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
Kirkland, J. L. & Tchkonia, T. Cellular senescence: a translational perspective. EBioMedicine 21, 21–28 (2017).
Niedernhofer, L. J. et al. Nuclear genomic instability and aging. Annu. Rev. Biochem. 87, 295–322 (2018).
Bussian, T. J. et al. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature 562, 578–582 (2018).
Camell, C. D. et al. Senolytics reduce coronavirus-related mortality in old mice. Science https://doi.org/10.1126/science.abe4832 (2021).
Chang, J. H. et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 22, 78–83 (2016).
Musi, N. et al. Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell https://doi.org/10.1111/acel.12840 (2018).
Ogrodnik, M. et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. https://doi.org/10.1038/ncomms15691 (2017).
Yousefzadeh, M. J. et al. An aged immune system drives senescence and ageing of solid organs. Nature 594, 100–105 (2021).
Zhang, P. S. et al. Senolytic therapy alleviates a beta-associated oligodendrocyte progenitor cell senescence and cognitive deficits in an Alzheimer’s disease model. Nat. Neurosci. 22, 719–728 (2019).
Kennedy, B. K. et al. Geroscience: linking aging to chronic disease. Cell 159, 708–712 (2014).
Basisty, N. et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. https://doi.org/10.1371/journal.pbio.3000599 (2020).
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Schafer, M. J. et al. The senescence-associated secretome as an indicator of age and medical risk. JCI Insight https://doi.org/10.1172/jci.insight.133668 (2020).
Ovadya, Y. et al. Impaired immune surveillance accelerates accumulation of senescent cells and aging. Nat. Commun. https://doi.org/10.1038/s41467-018-07825-3 (2018).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014).
Wiley, C. D. et al. SILAC analysis reveals increased secretion of hemostasis-related factors by senescent cells. Cell Rep. 28, 3329–3337 (2019).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Baker, D. J. et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
Zhu, Y. et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell 14, 644–658 (2015).
Wang, Y. Y. et al. Discovery of piperlongumine as a potential novel lead for the development of senolytic agents. Aging 8, 2915–2926 (2016).
Yousefzadeh, M. J. et al. Fisetin is a senotherapeutic that extends health and lifespan. EBioMedicine 36, 18–28 (2018).
Zhu, Y. et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 15, 428–435 (2016).
Baar, M. P. et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147 (2017).
He, Y. H. et al. Using proteolysis-targeting chimera technology to reduce navitoclax platelet toxicity and improve its senolytic activity. Nat. Commun. https://doi.org/10.1038/s41467-020-15838-0 (2020).
Amor, C. et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature 583, 127–132 (2020).
Childs, B. G. et al. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 (2017).
Niedernhofer, L. J. & Robbins, P. D. Senotherapeutics for healthy ageing. Nat. Rev. Drug Discov. 17, 377 (2018).
Hickson, L. J. et al. Senolytics decrease senescent cells in humans: preliminary report from a clinical trial of dasatinib plus quercetin in individuals with diabetic kidney disease. EBioMedicine 47, 446–456 (2019).
Justice, J. N. et al. Senolytics in idiopathic pulmonary fibrosis: results from a first-in-human, open-label, pilot study. EBioMedicine 40, 554–563 (2019).
Xu, M. et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med. 24, 1246–1256 (2018).
Milne, E. M. When does human ageing begin? Mech. Ageing Dev. 127, 290–297 (2006).
Yousefzadeh, M. J. et al. Tissue specificity of senescent cell accumulation during physiologic and accelerated aging of mice. Aging Cell 19, e13094 (2020).
Martin, G. M. Geroscience: addressing the mismatch between its exciting research opportunities, its economic imperative and its current funding crisis. Exp. Gerontol. 94, 46–51 (2017).
Hu, B. C. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019).
Saha, K. et al. The NIH somatic cell genome editing program. Nature 592, 195–204 (2021).
Dekker, J. et al. The 4D nucleome project. Nature 549, 219–226 (2017).
Consortium, G. T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Roy, A. L. et al. A blueprint for characterizing senescence. Cell 183, 1143–1146 (2020).
Parry, A. J. & Narita, M. Old cells, new tricks: chromatin structure in senescence. Mamm. Genome 27, 320–331 (2016).
Correia-Melo, C. et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 35, 724–742 (2016).
Liu, Y. et al. Expression of p16INK4a in peripheral blood T cells is a biomarker of human aging. Aging Cell 8, 439–448 (2009).
Wang, B. et al. An inducible p21-Cre mouse model to monitor and manipulate p21-highly-expressing senescent cells in vivo. Nat Aging 1, 962–973 (2021).
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995).
Freund, A., Laberge, R. M., Demaria, M. & Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23, 2066–2075 (2012).
Davalos, A. R. et al. p53-dependent release of Alarmin HMGB1 is a central mediator of senescent phenotypes. J. Cell Biol. 201, 613–629 (2013).
Hewitt, G. et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 3, 708 (2012).
Leontieva, O. V. & Blagosklonny, M. V. Gerosuppression in confluent cells. Aging 6, 1010–1018 (2014).
Severino, J., Allen, R. G., Balin, S., Balin, A. & Cristofalo, V. J. Is beta-galactosidase staining a marker of senescence in vitro and in vivo? Exp. Cell. Res. 257, 162–171 (2000).
Hall, B. M. et al. p16Ink4a and senescence-associated beta-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging 9, 1867–1884 (2017).
Hall, B. M. et al. Aging of mice is associated with p16Ink4a- and beta-galactosidase-positive macrophage accumulation that can be induced in young mice by senescent cells. Aging 8, 1294–1315 (2016).
Aix, E., Gutierrez-Gutierrez, O., Sanchez-Ferrer, C., Aguado, T. & Flores, I. Postnatal telomere dysfunction induces cardiomyocyte cell-cycle arrest through p21 activation. J. Cell Biol. 213, 571–583 (2016).
Puente, B. N. et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 157, 1243–1243 (2014).
Tane, S. et al. CDK inhibitors, p21Cip1 and p27Kip1, participate in cell cycle exit of mammalian cardiomyocytes. Biochem. Bioph. Res. Commun. 443, 1105–1109 (2014).
De Cecco, M. et al. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging 5, 867–883 (2013).
De Cecco, M. et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Campisi, J. et al. From discoveries in ageing research to therapeutics for healthy ageing. Nature 571, 183–192 (2019).
Sharpless, N. E. & Sherr, C. J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015).
Borner, K. et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 23, 1117–1128 (2021).
Acknowledgements
This research is supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under awards: U54AG075932 (to J.C. and B.S.), UG3CA268112 (to H.D.-L.), U54AG075934 (to L.D. and F.C.), UG3CA268117 (to Z.D.), U54AG076043 (to R.F. and S.H.), U54AG075931 (to T.F., M.K., A.L.M., O.E., M.R. and I.R.), UG3CA268096 (to L.G.), U54AG075941 (to G.K., V.G., N.M. and P.R.), UG3CA268091 (to J.H.L.), U54AG075936 (to P.J.L.), UG3CA268105 and U54AG075932 (to S.M.), UG3CA268202 (to N.N., S.W. and J.M.), U54AG076041 (to L.J.N. and C.F.A.), UG3CA268103 (to J.F.P.), U54AG076040 (to H.P.) and U24CA268108 (to J.C.S., Z.B.-J. and P.B.). We thank V. Bekker, SCENT Program Manager for administrative and formatting assistance. We also thank D. Mathias, Science Illustrator, for assistance with figures.
Author information
Authors and Affiliations
Consortia
Contributions
P.J.L., C.C.B., P.B., K.B., J.C., F.C., H.D.-L., P.D.J., L.D., F.E.D., O.E., R.F., T.F., D.F., V.G., N.G., C.G., I.H., Z.B.-J., P.K., S.K.,M.K., G.K., H.L., J.H.L., J.M., Q.M., S.M., K.M., A.L.M., N.M., N.N., J.F.P., I.R., J.C.R.-M., P.R., M.R., A.L.R., M.S.-K., B.S., P.S., J.C.S., V.S., J.X., J.W., A.I.W. and L.N wrote the manuscript; P.J.L, K.B., N.G., R.F. and S.K generated the figures; P.J.L., K.B., A.L.R., S.K., V.B., L.J.N., J.F.P. and R.F. reviewed and/or edited the manuscript; P.J.L., J.C., A.L.R., S.K. and L.J.N. contributed to discussion and provided critical review and/or revision of the manuscript. All other co-authors outside the writing group reviewed the manuscript and approved of its submission for publication.
Corresponding author
Ethics declarations
Competing interests
J.H.L. is an inventor on a pending patent applications related to Seq-Scope. L.G. is an inventor on two pending patent applications related to Pixel-seq. H.D.-L. has a research contract with MegaPro Biomedical and serves as managing director of a publishing company, Monasteria Press. R.F. is co-founder and scientific advisor of IsoPlexis, Singleron Biotechnologies and AtlasXomics. N.G. is a co-founder and equity owner of Datavisyn. J.C. receives research support from Ono, who are working on a new senolytic and have stock in Unity Biotechnology. The other authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Piero Carninci and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Fig. 1, methods and references
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
SenNet Consortium. NIH SenNet Consortium to map senescent cells throughout the human lifespan to understand physiological health. Nat Aging 2, 1090–1100 (2022). https://doi.org/10.1038/s43587-022-00326-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-022-00326-5
This article is cited by
-
Judith Campisi (1948–2024)
Nature Cell Biology (2024)
-
Lessons from inducible pluripotent stem cell models on neuronal senescence in aging and neurodegeneration
Nature Aging (2024)
-
MarsGT: Multi-omics analysis for rare population inference using single-cell graph transformer
Nature Communications (2024)
-
Telomeres, cellular senescence, and aging: past and future
Biogerontology (2024)
-
An open source knowledge graph ecosystem for the life sciences
Scientific Data (2024)